Short Biography
Nouria Hernandez obtained a Master (diploma) in Biology from the University of Geneva in 1980 and a PhD in Molecular Biology from the University of Heidelberg in 1983. She did postdoctoral research at Yale University, in New Haven, Connecticut, USA. In 1987, she joined the Cold Spring Harbor Laboratory, a research institute located on Long Island, New York, USA, first as a Senior Staff Investigator and then as a Senior Staff Scientist. In 1994, she was selected by the Howard Hughes Medical Institute, which then supported a large part of her research. In 2004, she spent a sabbatical at the University of Lausanne and joined the faculty there as a Professor and Director of the Center for Integrative Genomics in the fall of 2005. As of August 1st 2016, she has been the Rector of the University of Lausanne.

NOURIA HERNANDEZ – RESEARCH REPORT
Synthesis of non-coding RNAs by RNA polymerase III: mechanisms of transcription regulation
RNA polymerase III (pol III) is responsible for the transcription of a collection of short, non-coding RNAs involved in translation and a number of other essential cellular processes. For example, all tRNAs as well as the 5S RNA component of ribosomes are transcribed by pol III, and so is the essential RNA component of RNase P RNA, the enzyme involved in maturation of tRNAs. It has long been thought that all pol III genes are co-regulated, being for example transcribed at very high levels in exponentially growing cells and, conversely, at low levels in resting cells. However, measurements of pol III occupancy have revealed that many pol III genes, including tRNA genes, are in fact silent, even though their promoter sequences are intact.
A long standing interest has been to determine what causes a pol III gene to be either active or silent, and whether all pol III genes are indeed co-regulated. We have addressed these questions by measuring genome-wide pol III occupancy in human IMR90hTert cells under serum-replete or serum-starved conditions, as well as in serum-starved cells re-stimulated with insulin alone, or re-stimulated following addition of rapamycin, a specific inhibitor of the mTORC1 kinase. The mTORC1 pathway informs the cell about nutrient availability. Its inhibition under unfavorable conditions leads to the activation of the pol III repressor MAF1, which represses pol III transcription by binding to the enzyme itself and/or to its transcription factor TFIIIB. We performed chromatin immunoprecipitations with antibodies directed against a subunit of pol III followed by high throughput sequencing (ChIP-seq) experiments under these conditions, as well as in cells lacking MAF1. We observed a general pol III occupancy decrease in serum-starved conditions and an increase after insulin stimulation in the presence, but not in the absence, of the mTORC1 inhibitor rapamycin. This pol III occupancy decrease was, however, not uniform; rather, some highly pol III-occupied loci remained occupied even after serum starvation or rapamycin + insulin treatment, raising the possibility that these genes remained actively transcribed. Indeed, we excluded the possibility that the pol III signal detected on these genes corresponded to a “frozen” RNA polymerase that was not synthesizing RNA and could thus conclude that pol III repression under unfavorable conditions is actually not global: some genes, whose transcription is probably essential for cell survival in all conditions, remain highly transcribed.
To determine whether selective action of the repressor MAF1 might explain the escape of some pol III genes from repression, we measured MAF1 occupancy at pol III genes by the DamIP-seq method. We observed binding of MAF1 exclusively at pol III-occupied genes, and this binding increased under serum-starved conditions. MAF1 was also detectable at highly pol III-occupied genes whose pol III occupancy was not significantly decreased under serum starved conditions. The results revealed that MAF1 is present on pol III genes even under optimal growth conditions, suggesting that MAF1 acts in a chronic fashion, keeping pol III activity constantly in check, rather than in an acute fashion, repressing pol III transcription only under stress conditions. They also showed that selective escape of some pol III genes from repression could not be explained by selective MAF1 binding. We hypothesized that the negligible effect of MAF1 on some genes might result from their relatively high Pol III occupancy, as protein abundance studies by others in human cell lines estimate that MAF1 is 4-10 fold less abundant than most Pol III-specific subunits (Orioli et al., 2016, Genome Research 26, 624-635).
Another question we have been interested in is what are the consequences of deregulation of pol III transcription, specifically what are the consequences of lack of Maf1 in an organism such as a mouse. We continued our characterization of a mouse lacking the Maf1 gene in the entire body in collaboration with Dr. Ian Willis (Albert Einstein College of Medicine). Maf1 KO mice had a striking phenotype; they were lighter than the wild-type mice, had a dramatic reduction in white adipose tissue, and were slightly smaller than their wild-type counterparts. We performed a number of physiological as well as molecular studies to determine how lack of a pol III repressor might lead to this unexpected phenotype. We showed that Maf1 KO mice were unconditionally viable and resistant to diet-induced obesity and non-alcoholic fatty liver disease. Gene expression profiling of adipose tissue did not reveal significant changes in the transcriptome. Maf1 KO mice had, however, a reduced food intake and increased metabolic inefficiency, which did not result from induction of adaptive thermogenesis.
Maf1 being a pol III repressor, we determined pol III occupancy in wild-type and Maf1 KO mouse livers. We found increased pol III occupancy at pol III genes as well as increased levels of tRNA precursors. Unexpectedly, we did not observe increased levels of mature-length tRNAs, suggesting that one mechanism of increased energy expenditure in Maf1 KO mice is an increased synthesis and turnover (futile cycling) of tRNAs in the whole animal. We also showed futile cycling of hepatic lipids and reduced expression of nicotinamide N-methyltransferase (Nnmt) in liver and muscle. Lower levels of Nnmt have been shown to promote polyamine synthesis and enable nicotinamide salvage to regenerate NAD+ for oxidative metabolism. Indeed, metabolite profiling revealed elevated levels of many amino acids and spermidine. Maf1 KO mice displayed, moreover, an induction of autophagy and a slightly extended lifespan (Bonhoure et al. 2015, Genes & Development 29, 934-947). Thus, the absence of Maf1 in the entire animal leads to sever metabolic defects. A major interest is to link these effects to alteration of pol III products, in particular tRNAs.
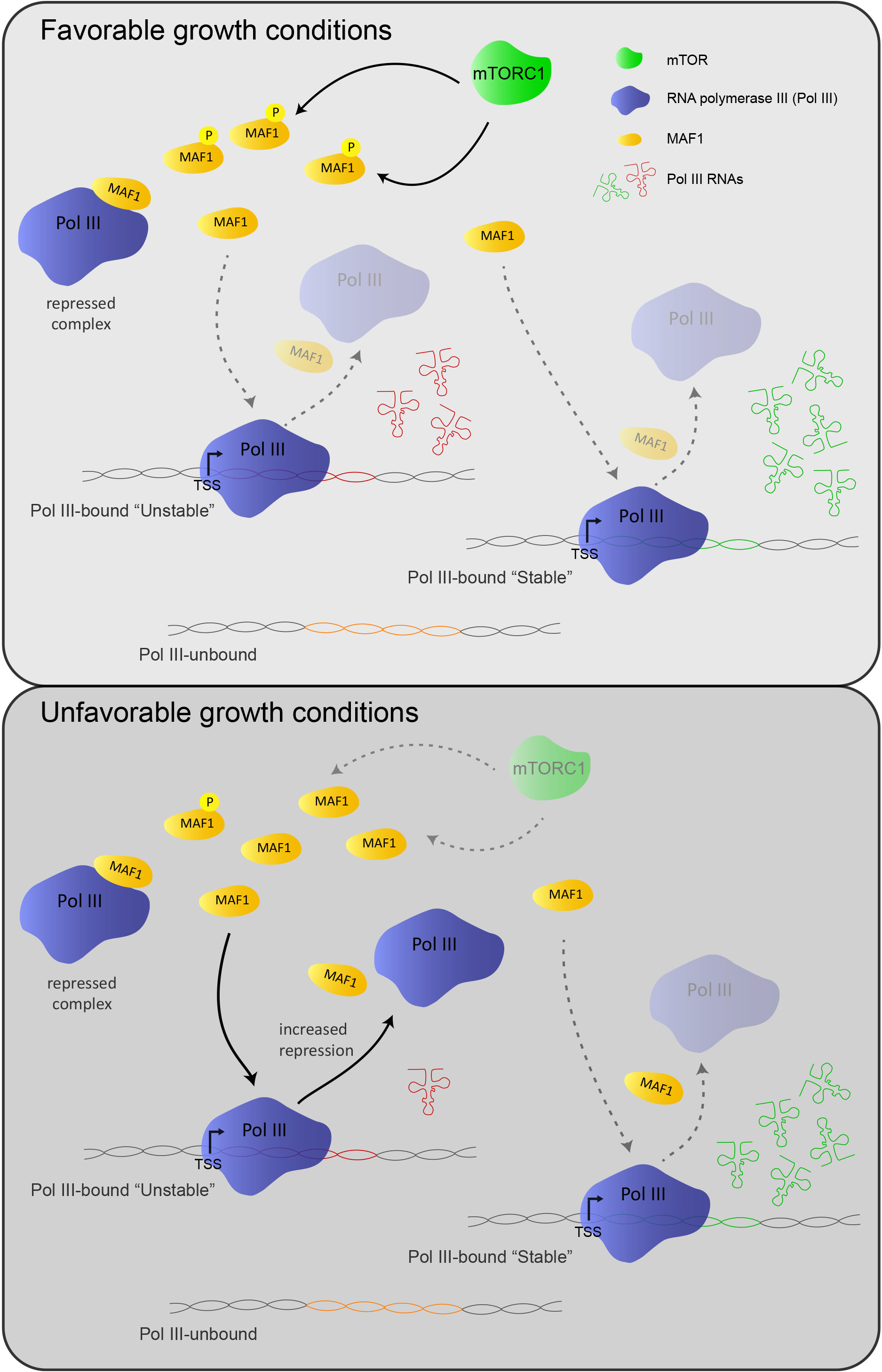
Fig 1. Model depicting MAF1-dependent regulation of Pol III transcription under different growth conditions. The tRNA molecules in red and green derive from unstable and stable genes, respectively (reproduced from Genome Research 2016, 26:624-635).
GROUP MEMBERS
Group Leader
Nouria Hernadez
nouria.hernandez@unil.ch
Bioinformatician
Viviane Praz
Postdoctoral Fellows
Oleksandr Dergai
Andrea Orioli
PhD Students
François Mange
Meghdad Yeganeh Seghalaksari
Master Student
Angela Etienne
Technicians
Pascal Cousin
Philippe Lhote
Gilles Willemin
Administrative assistant
Nathalie Clerc
nathalie.clerc@unil.ch
