Short Biography
David Gatfield received a diploma in biochemistry from the University of Tübingen (Germany), and a PhD for his work on mechanisms of mRNA transport and degradation with Elisa Izaurralde at the EMBL in Heidelberg. During his post-doctoral training with Ueli Schibler at the University of Geneva (Switzerland) he developed a dual interest in circadian clocks and RNA biology. He joined the CIG as a Swiss National Science Foundation professor in November 2010 and became associate professor in February 2017.

DAVID GATFIELD – RESEARCH REPORT
RNA Biology of circadian rhythms
Overview of our general aims and objectives
In mammals, most vital processes display circadian variations. Examples include sleep and wakefulness, body temperature, blood pressure, renal plasma flow, or the secretion of many hormones, which all show regular daily oscillations that persist even under constant conditions and that must thus be controlled by an endogenous pacemaker, known as the circadian clock.
Research in my group aims to uncover the molecular underpinnings of this circadian oscillator and of the pathways that it controls. According to the most commonly held molecular models of the mammalian circadian clock, the rhythm-generating core clock circuitry consists of an intricate network of transcription factors that interact in negative feedback loops; the rhythmic expression of core clock and of most clock-driven output genes would thus be engendered mainly at the transcriptional level.
In my lab, we focus on regulatory mechanisms that occur at the RNA level. These have been poorly explored so far and likely provide explanations to some of the enigmas in the chronobiology field. Conceivably, the post-transcriptional level also represents the ideal interface where DNA-encoded gene expression information and systemic or environmental signals – e.g. those emanating from feeding or body temperature rhythms – are integrated to yield the final, physiologically relevant output. With novel regulatory mechanisms and molecular players in hand, we aim to uncover their significance for daily physiology in vivo. Of note, it is our aim not only to contribute to a better understanding of chronobiology, but also to harness the circadian system as a paradigm for naturally occurring gene expression regulation in a key physiological process. With its different biological states just spaced by a few hours, the circadian system offers an exquisite experimental setting – amenable to research all the way from the behavioural to the molecular level – to study differential gene expression. The circadian field may hence ideally serve to uncover general principles of gene expression control and of RNA biology. To account for this dual interest, we have thus dubbed our research line “circadian RNA biology”.
Specific research lines
When setting up our first research activities at the CIG a few years back from now, we initially took a broad scope of circadian RNA biology that included projects on miRNAs, RNA-binding proteins, splicing, and translation. In the meanwhile, we have somewhat consolidated our research to two main interests, miRNAs and translation.
We aim to tackle two main scientific questions:
How do post-transcriptional mechanisms shape time-of-day-dependent transcriptomes and proteomes?
As mentioned above, the current molecular models assume that transcriptional oscillations drive the core clock and most clock-driven gene expression. However, this concept is at odds with observations that many rhythmic mRNAs do not originate from rhythmically transcribed genes, and that many rhythmic proteins are encoded by constantly abundant mRNAs. We have contributed to decipher parts of this enigma by analysing the role of miRNAs as drivers and modulators of rhythmic gene expression (Du, Arpat et al., eLife 2014) and by using a technique known as ribosome profiling to uncover rhythmicity at the translational level with high temporal and nucleotide resolution (Janich, Arpat et al., Genome Research 2015). In mouse liver, we identified ~150 mRNAs subject to robust daily changes in translational efficiency (see Movies 1 and 2). Rhythmic proteins produced from non-oscillating transcripts were associated with specific pathways, e.g. ribosome biogenesis and iron metabolism. With this inventory of rhythmically regulated protein biosynthesis in hand, we are now directing our efforts on gaining a better understanding of the molecular mechanisms, the signals, and the physiological functions of selected high- interest cases.
Several follow-up studies are emerging from these initial ribosome profiling-based endeavours. One of them aims to address how translational differences contribute to organ-specificity in gene expression oscillations (Castelo-Szekely et al., bioRxiv 2016). With this work we hope to be able to shed light on another poorly understood aspect of rhythmic gene expression: What are the mechanisms that make clock output rhythms so extraordinarily tissue-specific – despite the reliance on a core clock mechanism that appears to be largely shared between organs?
How does post-transcriptional control regulate the core clock circuitry?
Whether and how the core clock circuitry itself is subject to post-transcriptional control is largely unexplored. Conceivably, post-transcriptional control may be important to render core clock mRNAs sufficiently short-lived to warrant their rhythmic accumulation, or to modulate expression levels of individual core clock components, which in turn would impact on oscillation parameters such as phases and periods. We have, for example, found that miRNAs regulate the core clock component Per2 in liver and function in setting the period length of hepatic free-running circadian oscillations (Du, Arpat et al., eLife 2014). We are currently addressing further roles for miRNAs in influencing the kinetics of liver clock resetting by feeding, as well in the brain’s suprachiasmatic nucleus (i.e., in the mammalian central pacemaker). Moreover, our work on translation has uncovered several examples of core clock regulation. We are particularly excited about the identification of upstream open reading frame (uORF) translation as a novel regulatory mechanism within the clock circuitry (Janich, Arpat et al., Genome Research 2015; Castelo-Szekely et al., bioRxiv 2016). We have started work aimed at deciphering the functions of individual uORFs in core clock transcripts using genetic loss-of-function approaches in cells and in mice (Fig. 1).
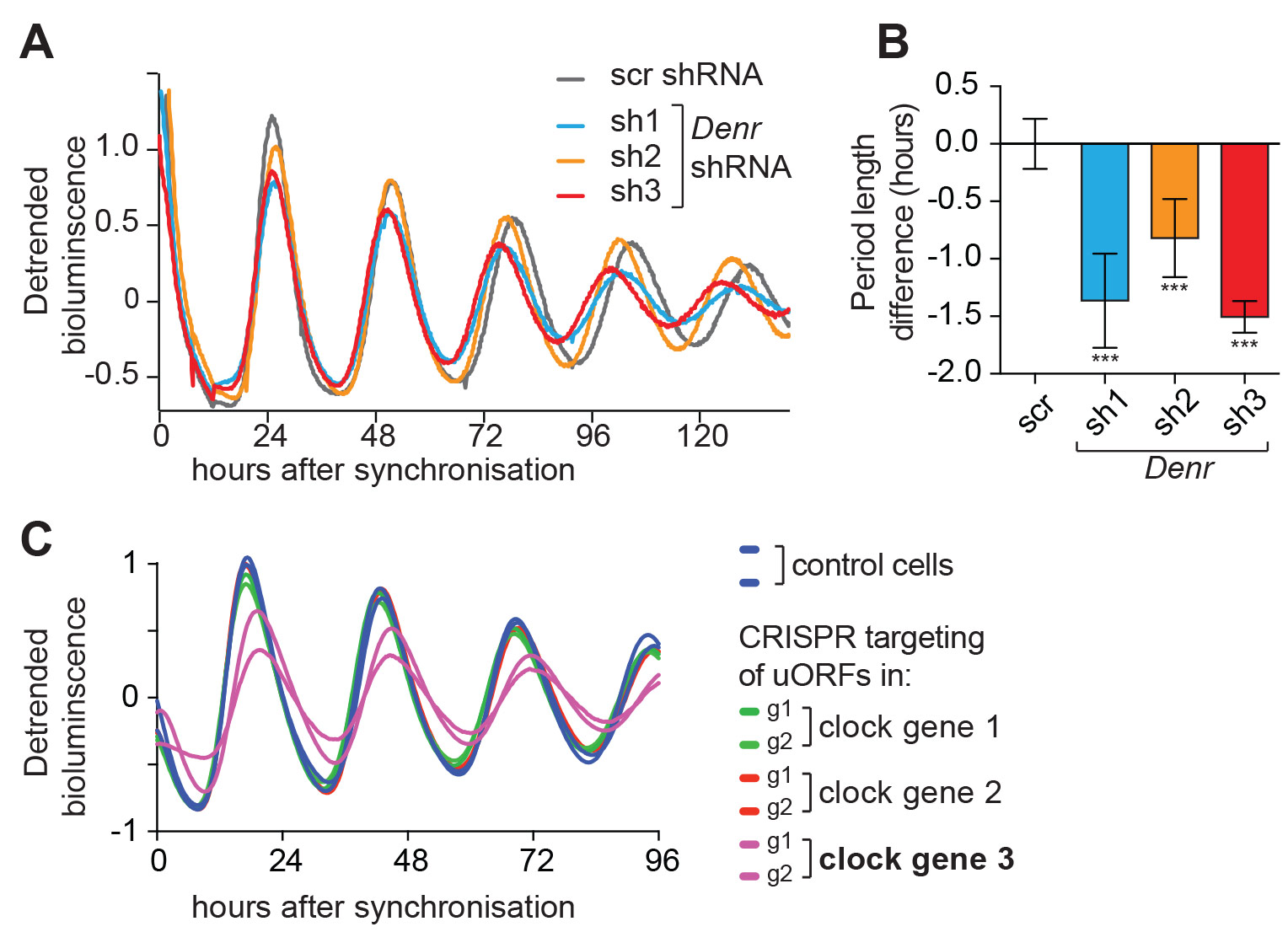
Fig 1. uORFs regulate the clock. A, B. Luciferase reporter rhythms measured in NIH3t3 cells that were depleted of a protein, DENR, that is necessary for translation re-initiation after uORF usage. As compared to control cells (scramble shRNA treated, scr), Denr knockdown with three difference shRNAs engenders short periods. (A) Shows representative raw data,
(B) a summary from several experiments. Data from Janich, Arpat et al., Genome Research, 2015.
(C.) Targeting the initiation codon of a specific uORF (in a gene termed “clock gene 3”, purple; two independent guide RNAs, g1 and g2) with CRISPR/Cas technology induces period lengthening, indicating that this particular uORF regulates clock parameters. (Unpublished data)
Movie 1. This movie shows an animation of the gene expression data from Janich, Arpat et al., Genome Research 2015, with transcript abundance on the abscissa and translation efficiency on the ordinate. Every dot corresponds to a single expressed gene in liver. Those ca. 150 transcripts for which specifically the translation efficiency, but not the transcript abundance, was detected as rhythmic, are shown in colour. Green colour shows the most prominent group of such transcripts, i.e. the mRNAs encoding ribosomal proteins, which all show a strong translational upsurge at around Zeitgeber Time (ZT) 10-12. This corresponds to the end of day/beginning of the night, when the nocturnal mice start becoming active and begin feeding. These rhythms are thus presumably feeding-dependent rather than driven by the cell-autonomous liver clock.
Movie 2. Similar to Movie 1, but this time all those transcripts were coloured, for which the rhythm already occurred at the mRNA abundance level (abscissa). For better visualisation, the colouring occurred in four different colours according to phases of maximal abundance.
GROUP MEMBERS
Group Leader
David Gatfield
david.gatfield@unil.ch
Bioinformatician
Bulak Arpat
Postdoctoral Fellows
Peggy Janich
Yannick Martinez
PhD Students
Violeta Castelo-Szekely
Ngoc-Hien Du
Technician
Mara De Matos
Technical Assistants
Sophie Debonneville
Maykel Duarte Lopes
Administrative Assistant
Carine Dovat
carine.dovat@unil.ch
