Short Biography
Vincent Dion first experienced laboratory research in 1999 as a summer student in Stanley L. Miller’s laboratory at UCSD (USA). He also spent time as an undergraduate in the laboratory of David H. Evans at the University of Guelph (Canada), where he got a B.Sc. in 2002. In 2007, he obtained his PhD from Baylor College of Medicine (USA) under the supervision of John H. Wilson, for defining the role of DNMT1, a DNA methyltransferase, in preventing disease-causing CAG/CTG repeat expansions. As a postdoc with Susan M. Gasser at the Friedrich Miescher Institute (Switzerland), he discovered a novel role for chromatin remodeling enzymes in the repair of deleterious DNA double-strand breaks. He joined the CIG in 2013 as an assistant professor with the support of a professorship from the Swiss National Science Foundation. Since then, his lab has made key contributions towards the development of gene editing approaches to reverse the expansion of CAG/CTG repeats.

VINCENT DION – RESEARCH REPORT
Chromatin, genome stability, and the expansion of trinucleotide repeats
Because we breathe oxygen and that most of our body is made out of water, our DNA is being damaged at a rate of about 100 000 lesions per cell per day. This enormous amount of damage must be repaired efficiently and accurately if the genetic information contained in the genome is to be maintained. This fundamental process of DNA repair is the focus of our laboratory.
Mutations in several DNA repair genes greatly increase cancer predisposition and the malfunction of several caretakers of the genome leads to neurological disorders. These observations highlight the importance of maintaining genome stability for human health.
Each cell in our body contains 2 meters worth of DNA that needs to be packaged into a nucleus of about 20 to 50 micron in diameter (a micron is 1/1’000’000 of a meter). To achieve this, DNA is wrapped around histone octamers to form nucleosomes and higher-order chromatin structures. Chromatin therefore helps to package the genome. Additionally, it provides great opportunities for the regulation of DNA-based events since nucleosomes can mask the binding site of many DNA binding proteins. This interplay between chromatin structure and DNA metabolism has been extensively studied in the context of transcription, but much less is known about its impact on DNA repair.
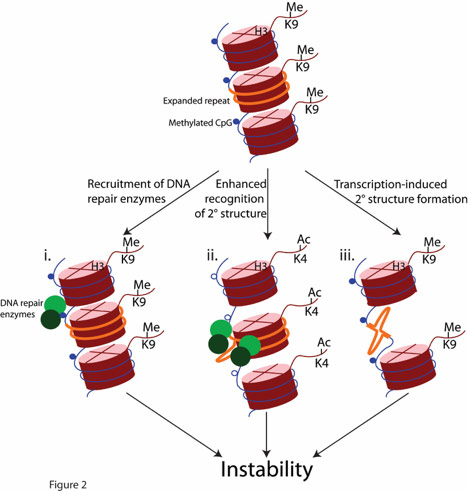
Fig 1. How chromatin structure could contribute to repeat instability (from Dion and Wilson Trends in Genetics 2009).
To study how chromatin affects DNA repair events, our laboratory focuses on tandem repeats composed of pure stretches of CAG or CTG trinucleotides. These loci are hotspots for genome instability and the size of the repeat can expand greatly, in some cases reaching thousands of trinucleotides at a single locus. As the repeat tract gets longer, it becomes more unstable and adopts chromatin configurations that bear many of the hallmarks of dense heterochromatin. The instability depends primarily on the normal activity of DNA repair on an unusual DNA sequence. These characteristics make trinucleotide repeats an ideal paradigm to explore the relationship between genome stability and chromatin structure at endogenous loci. We use a combination of cutting-edge microscopy, molecular biology, genetics, and genomics in cultured human cells to identify new players in trinucleotide repeat instability and to study the effects of chromatin structure and organization on DNA repair. Specifically, we are trying to answer the following questions:
1) Do chromatin modifiers affect repeat instability directly by changing the local structure of the expanded repeat tract? Many chromatin modifying enzymes have been implicated in repeat instability but their exact role is unclear. This is because every study conducted so far includes loss of function experiments that do not permit the differentiation between direct or indirect causes, such as changes in genome-wide transcription. Here we are designing novel molecular assays to address the roles of chromatin modifying enzymes in repeat instability.
2) What is the impact of nuclear organization on trinucleotide repeat instability? Here we aim to determine whether expanded trinucleotide repeats impact their 3D organization in the nucleus and whether this organization in turn influences their repair and thus their instability.
3) Can we induce CAG repeat contractions without also inducing expansions? This project is especially relevant to human health. Indeed, the expansion of trinucleotide repeats causes a number of neurological and neuromuscular disorders, including Huntington disease, myotonic dystrophy, Fragile X syndrome, as well as several spinocerebellar ataxias. All of these disorders remain without a cure. This would, in theory, remove the underlying cause of the disease and provide a therapeutic avenue. This project, therefore, is focused on finding conditions or treatments that specifically contract the repeat tracts and that would therefore alleviate the molecular symptoms. We have recently shown that the CRISPR-Cas9 nickase can be targeted to the expanded repeat tract and induces almost exclusively contractions (Cinesi et al., Nat Comms 2016), which is an exciting first step towards the development of gene editing approaches for therapeutic purposes.
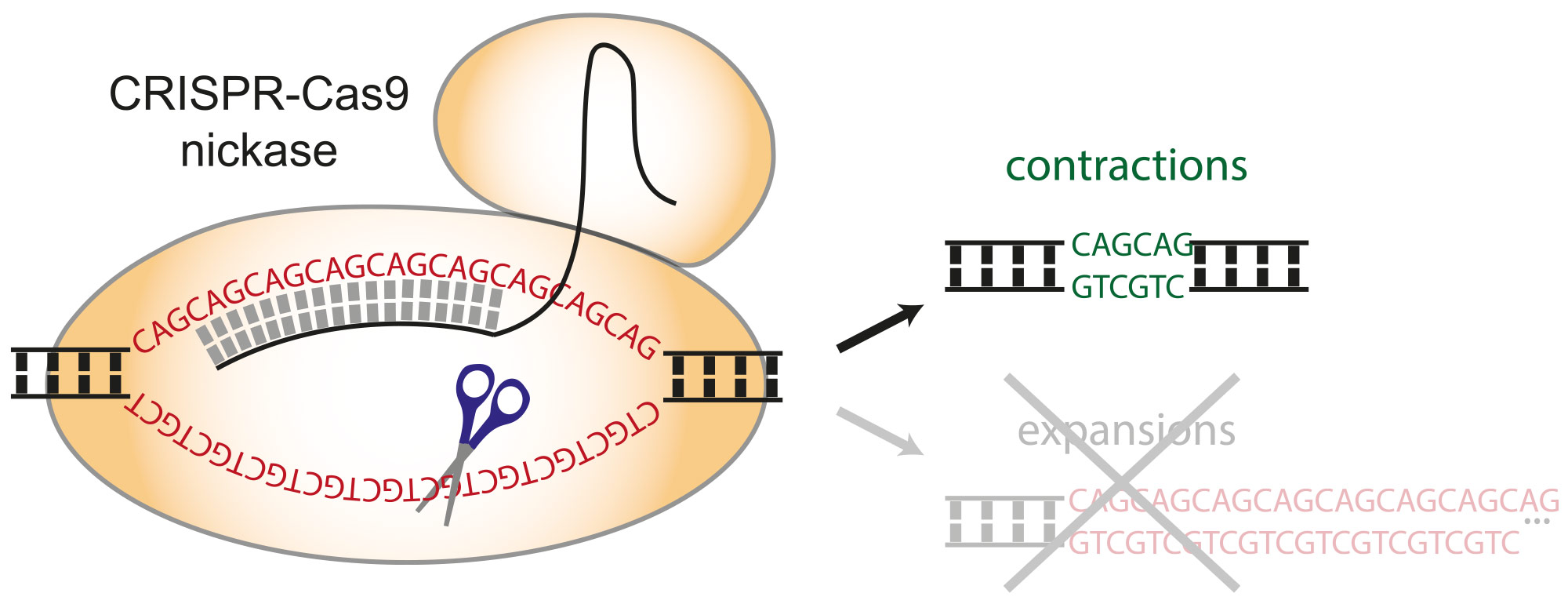
Fig 2. The CRISPR-Cas9 nickase can be directed to an expanded CAG/CTG repeat tract using a guide RNA that targets it specifically. That leads to cutting of one of the two DNA strand. At expanded repeats, this leads to the formation of several nicks on the same strand, which creates a single-stranded DNA gap that leads to repeat contraction without also provoking repeat expansions. This is the first tool for gene editing that induces contractions specifically, thereby correcting the disease causing mutation.
A long term goal of our research is to understand the mechanism of trinucleotide repeat instability such that we can manipulate it at will and induce repeat contractions in patients. We are especially interested in being able to manipulate local chromatin structure to change the bias in repeat expansion usually seen in human patients.
GROUP MEMBERS
Group Leader
Vincent Dion
vincent.dion@unil.ch
Postdoctoral Fellows
Flavia Marzetta
Oscar Rodriguez Lima
PhD Students
Cinzia Cinesiv
Gustavo A. Ruiz Buendia
Bin Yang
Master Students
Melvin Berard
Alicia Borgeaud
Nastassia Gobet
Mélanie Op
Evgeniya Trofimenko
Paul Zganiacz
Technician
Lorène Aeschbach
Fisrt-Step Student
Jean-Jerrold Pierre
Summer Student
Johanna Elizabeth Martin
Administrative assistant
Nathalie Clerc
nathalie.clerc@unil.ch
